Abstract
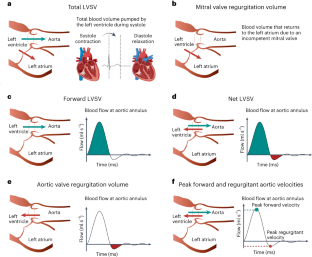
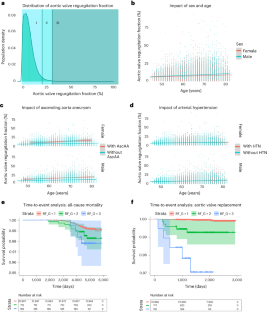
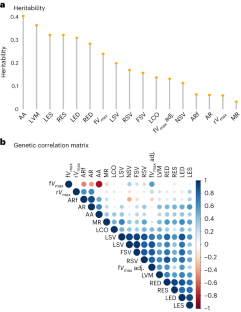
Cardiac blood flow is a critical determinant of human health. However, the definition of its genetic architecture is limited by the technical challenge of capturing dynamic flow volumes from cardiac imaging at scale. We present DeepFlow, a deep-learning system to extract cardiac flow and volumes from phase-contrast cardiac magnetic resonance imaging. A mixed-linear model applied to 37,653 individuals from the UK Biobank reveals genome-wide significant associations across cardiac dynamic flow volumes spanning from aortic forward velocity to aortic regurgitation fraction. Mendelian randomization reveals a causal role for aortic root size in aortic valve regurgitation. Among the most significant contributing variants, localizing genes (near ELN, PRDM6 and ADAMTS7) are implicated in connective tissue and blood pressure pathways. Here we show that DeepFlow cardiac flow phenotyping at scale, combined with genotyping data, reinforces the contribution of connective tissue genes, blood pressure and root size to aortic valve function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout

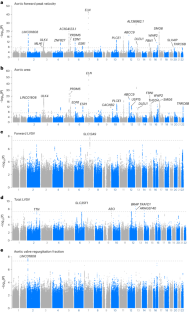
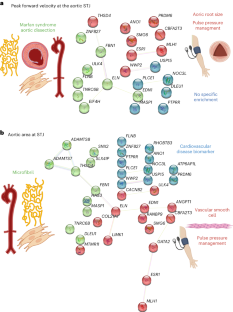
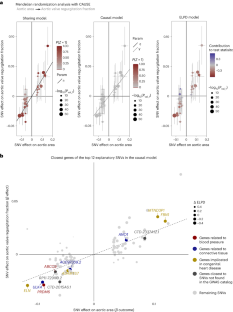
Similar content being viewed by others
Data availability
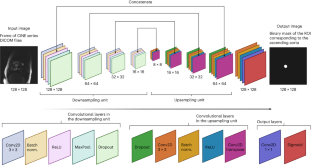
UK Biobank data is accessible to researchers with genuine research purposes, subject to institutional review board and UK Biobank approval (http://www.ukbiobank.ac.uk/register-apply/). The GWAS summary statistics can be obtained from Zenodo at https://zenodo.org/record/8388416. The Genotype-Tissue Expression v.8 datasets can be found at https://www.gtexportal.org/home/datasets. The STRING database can be accessed at https://string-db.org/. The graphical representations of the Mask-R-CNN and the FPN model architectures can be accessed from Zenodo at https://doi.org/10.5281/zenodo.8384058. Any additional data is either within the article or its supplementary materials. Source data are provided with this paper.
Code availability
The custom software, including model segmentation training and validation, is freely available for non-commercial use (open-source CC-BY-NC license) at https://github.com/Urban90/deepFlow and https://zenodo.org/badge/latestdoi/487159922 (ref. 76).
References
-
Virani, S. S. et al. Heart disease and stroke statistics-2021 update: a report from the American Heart Association. Circulation 143, e254–e743 (2021).
-
Nauffal, V. et al. Genetics of myocardial interstitial fibrosis in the human heart and association with disease. Nat. Genet. 55, 777–786 (2023).
-
Thanaj, M. et al. Genetic and environmental determinants of diastolic heart function. Nat. Cardiovasc. Res. 1, 361–371 (2022).
-
Bai, W. et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 20, 65 (2018).
-
Pirruccello, J. P. et al. Deep learning of left atrial structure and function provides link to atrial fibrillation risk. Preprint at medRxiv 2021.08.02.21261481 (2021).
-
Davies, R. H. et al. Precision measurement of cardiac structure and function in cardiovascular magnetic resonance using machine learning. J. Cardiovasc Magn. Reson. 24, 16 (2022).
-
Nayak et al. Cardiovascular magnetic resonance phase contrast imaging. J. Cardiovasc Magn. Reson. 17, 71 (2015).
-
Malhotra, P., Gupta, S., Koundal, D., Zaguia, A. & Enbeyle, W. Deep neural networks for medical image segmentation. J. Health. Eng. 2022, 9580991 (2022).
-
Aung, N. et al. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci associated with cardiac morphogenesis and heart failure development. Circulation 140, 1318–1330 (2019).
-
Petersen, S. E. et al. UK Biobank’s cardiovascular magnetic resonance protocol. J. Cardiovasc. Magn. Reson. 18, 8 (2016).
-
Garg, P. et al. Assessment of mitral valve regurgitation by cardiovascular magnetic resonance imaging. Nat. Rev. Cardiol. 17, 298–312 (2020).
-
Benjamins, J. W. et al. Genomic insights in ascending aortic size and distensibility. EBioMedicine. 75, 103783 (2022).
-
Heiberg, E. et al. Design and validation of segment-freely available software for cardiovascular image analysis. BMC Med. Imaging 10, 1 (2010).
-
Bekeredjian, R. & Grayburn, P. A. Valvular heart disease: aortic regurgitation. Circulation 112, 125–134 (2005).
-
DesJardin, J. T., Chikwe, J., Hahn, R. T., Hung, J. W. & Delling, F. N. Sex differences and similarities in valvular heart disease. Circ. Res. 130, 455–473 (2022).
-
Nitsche, C., Koschutnik, M., Kammerlander, A., Hengstenberg, C. & Mascherbauer, J. Gender-specific differences in valvular heart disease. Wien. Klin. Wochenschr. 132, 61–68 (2020).
-
Bonow, R. O. et al. Focused update incorporated into the ACC/AHA 2006 guidelines for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Revise the 1998 Guidelines for the Management of Patients With Valvular Heart Disease): endorsed by the Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. Circulation 118, e523–e661 (2008).
-
Córdova-Palomera, A. et al. Cardiac imaging of aortic valve area from 34 287 UK Biobank participants reveals novel genetic associations and shared genetic comorbidity with multiple disease phenotypes. Circ. Genom. Precis. Med. 13, e003014 (2020).
-
Spampinato, R. A. et al. Grading of aortic regurgitation by cardiovascular magnetic resonance and pulsed Doppler of the left subclavian artery: harmonizing grading scales between imaging modalities. Int. J. Cardiovasc. Imaging 36, 1517–1526 (2020).
-
Myerson, S. G. et al. Aortic regurgitation quantification using cardiovascular magnetic resonance: association with clinical outcome. Circulation 126, 1452–1460 (2012).
-
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
-
Eijgelsheim, M. et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum. Mol. Genet. 19, 3885–3894 (2010).
-
Lin, H. et al. Common and rare coding genetic variation underlying the electrocardiographic PR interval. Circ. Genom. Precis. Med. 11, e002037 (2018).
-
Derks, W. & Bergmann, O. BRAP: a novel regulator of the cardiomyocyte cell cycle controlling both proliferation and survival? Cardiovasc. Res. 116, 467–469 (2020).
-
Volland, C. et al. Control of p21Cip by BRCA1-associated protein is critical for cardiomyocyte cell cycle progression and survival. Cardiovasc. Res. 116, 592–604 (2020).
-
Wain, L. V. et al. Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension 70, e4–e19 (2017).
-
Verweij, N. et al. The genetic makeup of the electrocardiogram. Cell Syst. 11, 229–238 (2020).
-
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
-
Welter, D. et al. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
-
Surendran, P. et al. Discovery of rare variants associated with blood pressure regulation through meta-analysis of 1.3 million individuals. Nat. Genet. 52, 1314–1332 (2020).
-
Van der Harst, P. & Verweij, N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 122, 433–443 (2018).
-
Ishigaki, K. et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 52, 669–679 (2020).
-
Hoffmann, T. J. et al. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat. Genet. 49, 54–64 (2017).
-
Verweij, N., van de Vegte, Y. J. & van der Harst, P. Genetic study links components of the autonomous nervous system to heart-rate profile during exercise. Nat. Commun. 9, 898 (2018).
-
Ramírez, J. et al. Thirty loci identified for heart rate response to exercise and recovery implicate autonomic nervous system. Nat. Commun. 9, 1947 (2018).
-
Saw, J. et al. Chromosome 1q21.2 and additional loci influence risk of spontaneous coronary artery dissection and myocardial infarction. Nat. Commun. 11, 4432 (2020).
-
Müller, R. et al. ANGIOGENES: knowledge database for protein-coding and noncoding RNA genes in endothelial cells. Sci. Rep. 6, 32475 (2016).
-
Francis, C. M. et al. Genome-wide associations of aortic distensibility suggest causality for aortic aneurysms and brain white matter hyperintensities. Nat. Commun. 13, 4505 (2022).
-
Svendsen, J. M. et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138, 63–77 (2009).
-
Snow, B. E. et al. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 13, 698–704 (2003).
-
Chakravarti, S., Enzo, E., de Barros, M. R. M., Maffezzoni, M. B. R. & Pellegrini, G. Genetic disorders of the extracellular matrix: from cell and gene therapy to future applications in regenerative medicine. Annu. Rev. Genomics Hum. Genet. 23, 193–222 (2022).
-
Chai, T. et al. Genome-wide identification of RNA modifications for spontaneous coronary aortic dissection. Front. Genet. 12, 696562 (2021).
-
Wang, Y. et al. EPHB4 protein expression in vascular smooth muscle cells regulates their contractility, and EPHB4 deletion leads to hypotension in mice. J. Biol. Chem. 290, 14235–14244 (2015).
-
Temprano-Sagrera, G. et al. Multi-phenotype analyses of hemostatic traits with cardiovascular events reveal novel genetic associations. J. Thromb. Haemost. 20, 1331–1349 (2022).
-
Jeong, H., Jin, H. S., Kim, S. S. & Shin, D. Identifying interactions between dietary sodium, potassium, sodium–potassium ratios, and FGF5 rs16998073 variants and their associated risk for hypertension in Korean adults. Nutrients 12, 2121 (2020).
-
Szklarczyk, D. et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
-
Pers, T. H. et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 6, 5890 (2015).
-
Abecasis, G. R. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
-
Morrison, J., Knoblauch, N., Marcus, J. H., Stephens, M. & He, X. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat. Genet. 52, 740–747 (2020).
-
Vehtari, A., Gelman, A. & Gabry, J. Practical Bayesian model evaluation using leave-one-out cross-validation and WAIC. Stat. Comput. 27, 1413–1432 (2017).
-
Bethesda (MD): National Library of Medicine (US), N.C.f.B.I. Gene (ULK4). https://www.ncbi.nlm.nih.gov/gene/54986
-
Silva, C. T. et al. A combined linkage and exome sequencing analysis for electrocardiogram parameters in the Erasmus Rucphen family study. Front. Genet. 7, 190 (2016).
-
Ntalla, I. et al. Multi-ancestry GWAS of the electrocardiographic PR interval identifies 202 loci underlying cardiac conduction. Nat. Commun. 11, 2542 (2020).
-
Malfait, F., Wenstrup, R. J. & De Paepe, A. Clinical and genetic aspects of Ehlers-Danlos syndrome, classic type. Genet. Med. 12, 597–605 (2010).
-
Jones, J. A. & Ikonomidis, J. S. The pathogenesis of aortopathy in Marfan syndrome and related diseases. Curr. Cardiol. Rep. 12, 99–107 (2010).
-
Datta, A. S., Zhang, Y., Zhang, L. & Biswas, S. Association of rare haplotypes on ULK4 and MAP4 genes with hypertension. BMC Proc. 10, 363–369 (2016).
-
Guo, D. C. et al. Genetic variants in LRP1 and ULK4 are associated with acute aortic dissections. Am. J. Hum. Genet. 99, 762–769 (2016).
-
Pirruccello, J. P. et al. Deep learning enables genetic analysis of the human thoracic aorta. Nat. Genet. 54, 40–51 (2022).
-
Matchkov, V. V. et al. A paradoxical increase of force development in saphenous and tail arteries from heterozygous ANO1 knockout mice. Physiol. Rep. 8, e14645 (2020).
-
Liu, C. J. et al. ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 20, 988–990 (2006).
-
Bauer, R. C. et al. Knockout of Adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation 131, 1202–1213 (2015).
-
Erbel, R. et al. ESC guidelines on the diagnosis and treatment of aortic diseases: document covering acute and chronic aortic diseases of the thoracic and abdominal aorta of the adult. The Task Force for the Diagnosis and Treatment of Aortic Diseases of the European Society of Cardiology (ESC). Eur. Heart J. 35, 2873–2926 (2014).
-
O’Sullivan, J. W., Ashley, E. A. & Elliott, P. M. Polygenic risk scores for the prediction of cardiometabolic disease. Eur. Heart J. 44, 89–99 (2023).
-
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
-
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
-
Lin, T.-Y. et al. Feature pyramid networks for object detection. Preprint at arXiv https://doi.org/10.48550/arXiv.1612.03144 (2017).
-
He, K., Gkioxari, G., Dollár, P. & Girshick, R. Mask R-CNN. IEEE Trans. Pattern Anal. Mach. Intell. 42, 386–397 (2020).
-
Zhou, W. et al. Efficiently controlling for case–control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018).
-
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
-
Shah, S. et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 11, 163 (2020).
-
Lin, Z., Knutson, K. A. & Pan, W. Leveraging omics data to boost the power of genome-wide association studies. HGG Adv. 3, 100144 (2022).
-
Wallace, C. Statistical testing of shared genetic control for potentially related traits. Genet. Epidemiol. 37, 802–813 (2013).
-
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
-
Köhler, S. et al. The human phenotype ontology in 2021. Nucleic Acids Res. 49, D1207–D1217 (2021).
-
Davey Smith, G. & Hemani, G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–R98 (2014).
-
Singh, A. & Gomes, B. Urban90/deepFlow: v1.0.0—initial stable release. Zenodo https://doi.org/10.5281/zenodo.8387683 (2023).
Acknowledgements
This research has been conducted using the UK Biobank Resource under applications 63735 and 22282. Figures were created with BioRender.com and LocusZoom.org. We acknowledge the following financial support: Deutsche Forschungsgemeinschaft Walter-Benjamin Program GO 3196/3-1, 707766—809341 (to B.G.); Leducq Foundation Transatlantic Network of Excellence 21CVD02 (to B.G., B.M., and E.A.); GREGoR U01HG011762 and R01HL142015 (to P.C.G.); Novo Nordisk Foundation NNF19OC0054265 (to T.M.S.); National Medical Research Council Singapore NMRC-IRG A-0006207-00-00 (to R.F.) and National University of Singapore NUS-MSRMP 2023 (to S.L.).
Author information
Authors and Affiliations
Contributions
B.G. and E.A. designed the study. B.G. trained, validated and tested DeepFlow with contribution of A.S. and M.K.; conducted the GWAS and epidemiology-related analyses, prepared the supplementary material and tables; designed the figures with the contribution of A.S. and S.L. B.G., E.A. and V.P. wrote the manuscript with contributions of A.S., D.A., J.S., M.S., S.L., R.F. and F.H. B.M. gave advice in the conception of the project. A.S. performed software implementation of DeepFlow, as well as ensured cross-platform compatibility. A.S. contributed to the time-to-event analyses of clinical outcomes in aortic valve regurgitation. D.A. contributed and advised on the Mendelian randomization analysis. T.M.S., P.C.G., S.B.M., S.L. and R.F. were instrumental in the revision of this work. P.C.G. performed the eQTL colocalization analysis. J.W.H. advised regarding the benchmarking against other deep-learning-based image segmentation models.
Corresponding author
Ethics declarations
Competing interests
B.M. is part of the Scientific Advisory Board of Fleischhacker GmbH. E.A. is the founder of Personalis, DeepCell, Svexa, Overtone and Parameter Health; advisor to Pacific Biosciences, SequenceBio, Nuevocor and Apple and a nonexecutive director of AstraZeneca. J.S. is a consultant for Google AI. S.B.M. is an advisor to BioMarin, Myome and Tenaya Therapeutics. V.P. is SAB for BioMarin and Lexeo Therapeutics; a consultant for Viz.ai and Nuevocor and has sponsored research from Biomarin, Inc. and Saliogen Therapeutics. M.S. receives research support from Siemens Healthineers and GE Healthcare and is a consultant to Medtronic, G3 and Imagion Biosciences. All other authors have no conflicts of interest to declare.
Peer review
Peer review information
Nature Genetics thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–7, Supplementary Figs. 1–12, Supplementary Notes 1–15 and Supplementary Equations.
Supplementary Data 1
Annotations and cross-references with GWAS catalog for the independently associated SNVs, as well as genomic risk loci definitions with 12 tab-separated tables including six tables with annotated independently significantly and nominally associated SNVs across phenotypes (filename starting with ‘IndSigSNPs_annotated’; six tables defining the significantly and nominally associated genomic risk loci across phenotypes (filename starting with GenomicRiskLoci). Phenotype names are given in the filename: ‘iaorta_area’ for BSA-indexed aortic area; ‘vmax’ for peak forward aortic velocity; ‘tLVSV’ for BSA-indexed total LVSV; ‘fLVSV’ for BSA-indexed forward LVSV; ‘vmax_adjusted’: peak forward aortic velocity adjusted to the aortic area; ‘ao_regurgitation_fraction’: aortic valve regurgitation fraction for the annotated SNV tables, the columns denote: ‘GenomicLocus’:in which genomic risk locus the SNV is located (each genomic risk locus is assigned a number, shown in the GenomicRiskLoci tables); ‘IndSignSNP_GWAS’: the SNV id following the format chr:pos:affected allele:another allele in hg19, or rsid if available; ‘P value GWAS’: the respective P value in the GWAS; ‘nearestGene’: the nearest gene; ‘CADD’ the CADD score; ‘RDB’: Regulome DB score; ‘snp_PMID’: the SNV rsid described in the literature; ‘chr’: chromosome of the SNV described in the literature; ‘bp’: base pair position of the SNV described in the literature; ‘PMID’: PubMed id; ‘Study’: study name; ‘Trait’: phenotype studied in the GWAS reported in the literature; ‘ReportedGene’: reported gene associated with the SNV in the literature; ‘DateAddedToCatalog’: date the GWAS from the literature was added to the GWAS catalog; ‘P value_PMID’: calculated P value for the SNV reported in the literature. For the genomic risk loci tables: ‘GenomicLocus’: index of the genomic risk locus; ‘Novely’: indicated by an asterisk symbol, if the genomic risk locus is considered novel: either at the genomic level, if associations were found for traits not identified in previous GWAS analyses of comparable traits (aortic area defined in this study versus aortic functional/morphology traits; total LVSV versus left ventricular volumes); or at the phenotypic level, associations were observed for previously unstudied traits; ‘uniqID’: Unique ID of SNVs consisting of chr:position:reference allele:alternative allele; ‘rsID’: rsID of the top lead SNV based on dbSNP build 146; ‘chr’: chromosome of top lead SNV; ‘pos’: position of top lead SNV on hg19; ‘p’: P value of top lead SNV; ‘start’: start position of the locus; ‘end’: end position of the locus; nSNVs: the number of unique candidate SNVs in the genomic locus, including non-GWAS-tagged SNPs (which are available in the 1KG/Phase 3 reference panel. Candidate SNPs are all SNVs that are in LD (r2 < 0.6) with any of independent significant SNVs and either have a P value below the threshold for nominal significance or are only available in 1,000G; ‘nGWASSNPs’: The number of unique GWAS-tagged candidate SNVs in the genomic locus which is available in the GWAS summary statistics input file, which is a subset of ‘nSNPs’; ‘nIndSigSNPs’: the number of the independent (r2 < 0.6) significant SNVs in the genomic locus; ‘IndSigSNPs’: rsID of the independent significant SNVs in the genomic locus; ‘nLeadSNPs’: the number of lead SNVs in the genomic locus, lead SNVs are subset of independent significant SNVs at r2 of 0.1; ‘LeadSNPs’: rsID of lead SNVs in the genomic locus.
Supplementary Data 2
eQTL colocalization results. Table with the eQTL colocalization results across phenotypes with at least one nominally significantly associated SNV. Phenotypes: ‘iaorta_area’: BSA-indexed aortic area; ‘Vmax’: peak forward aortic velocity; ‘tLVSV’: BSA-indexed total LVSV; ‘fLVSV’: BSA-indexed forward LVSV. ‘sentinel’: sentinel GWAS variants (with the strongest association) which were at least 1 megabase apart; ‘gwas’: phenotype analyzed in the GWAS; ‘qtl’ tissue type of the GTEx eQTLs version 8; ‘pval_gwas’: associated P value in the GWAS; ‘pval_eqtl’: GTEx P value of the top significant eQTL variant for the given gene in the given tissue within 10 kb of the GWAS variant; ‘identifier’: gene identifier; ‘gene_name’: name of the gene; ‘gene_biotype’: classification of a gene based on the type of transcript it produces and its associated function; ‘gene_description’: description of the gene; ‘COLOC_n_snps’: number of variants in this region (1 Mb window around the sentinel GWAS variant) that were present in both the GWAS and eQTL data and thus contributed to the colocalization estimate; ‘COLOC_h4’: posterior probability that both the gene eQTL and the trait of interest are driven by the same causal variant.
Supplementary Data 3
Pathway enrichment analysis results with STRING. Six tab-separated tables corresponding to the pathway enrichment analysis clusters (n = 3: ‘blue’, ‘green’ and ‘red’ clusters) per phenotype (BSA-indexed aortic area—‘iaorta_area’; and peak forward aortic velocity—‘Vmax’). ‘#category’: database used in the STRING analysis; ‘term ID’: unique identifier assigned to a term in a controlled vocabulary or ontology; ‘term description’: brief definition of the meaning and scope of the term id; ‘observed gene count’: number of genes included for analysis from GWAS results; ‘background gene count’: number of proteins in the reference genome (PMID 33237311); ‘strength’: strength of an interaction, which is represented by a numeric value between 0 and 1, where 1 indicates the highest level of confidence and 0 indicates the lowest level of confidence, moreover the strength score reflects the degree of experimental support, co-expression correlation, text mining, that support the interaction between the two genes/proteins; ‘FDR’: false discovery rate; ‘matching proteins in your network (IDs)’: identifiers of the gene/proteins in the network; ‘matching proteins in your network (labels)’: names of the genes/proteins in the network.
Supplementary Data 4
Tissue enrichment results using the DEPICT software. Four tab-separated files with the tissue enrichment results using the DEPICT software for the phenotypes aortic area (‘iaorta_area’), aortic peak forward velocity (‘Vmax’), aortic regurgitation fraction (‘regurgitation_fraction’) and aortic peak forward velocity adjusted to aortic area (‘Vmax_adjusted’). The column names indicate as defined by the DEPICT software: ‘MeSH term’: (Medical Subject Heading; http://www.nlm.nih.gov/mesh/) term for the tissue or cell type annotation; ‘name’: name of the tissue or cell type; ‘MeSH first level term’: description of the tissue or cell type annotation; ‘MeSH second level term’: more general description of the tissue or cell type annotation; ‘nominal P value’: nominal enrichment P value of tissue/cell type annotation (null hypothesis: genes in associated are not highly expressed in the given tissue or cell type): ‘FDR’: estimated FDR for the tissue or cell type; ‘tissue-specific expression z score gene’: genes highly expressed in tissue/cell type and overlapping with associated loci. The z score denotes the level of tissue-specific expression.
Source data
Source Data Fig. 5
Statistical source data.
Source Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gomes, B., Singh, A., O’Sullivan, J.W. et al. Genetic architecture of cardiac dynamic flow volumes. Nat Genet (2023). https://ift.tt/zFTRnN6
-
Received:
-
Accepted:
-
Published:
-
DOI: https://ift.tt/zFTRnN6
"flow" - Google News
December 11, 2023 at 05:13PM
https://ift.tt/B3l0G5I
Genetic architecture of cardiac dynamic flow volumes - Nature.com
"flow" - Google News
https://ift.tt/yPMD9u8
https://ift.tt/KhfNMu2
Bagikan Berita Ini















0 Response to "Genetic architecture of cardiac dynamic flow volumes - Nature.com"
Post a Comment